Importing Data
📦 Learn how to import raw data into Isabl using existing metadata.
Isabl-CLI enables tracking and managing of raw data, as well as reference resources that are a function of a genome assembly or an experimental technique.
Data Import
Isabl-CLI supports automated data import by recursively exploring data deposition directories and matching raw data files with identifiers registered in the database. For example, the client can be instructed to explore the /projects directory (A), retrieving only samples from Project 393, and match files using Sample Identifiers (B).
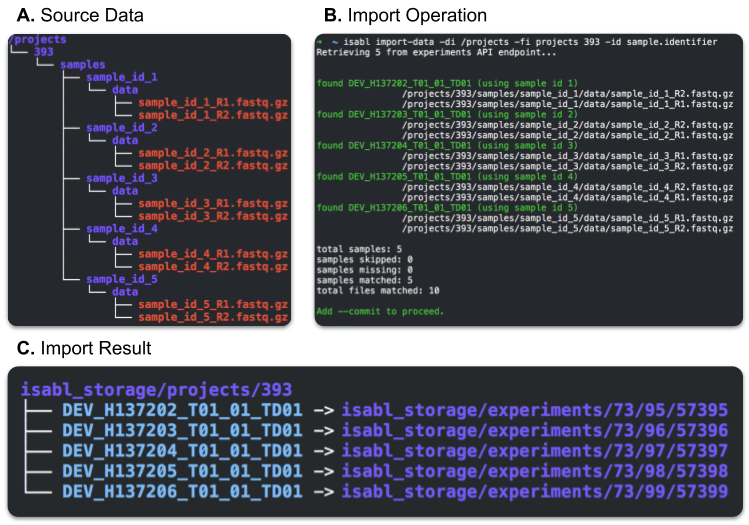
Upon --commit, Isabl-CLI proceeds to move (or symlink) matched files into scalable directory structures (C). The experiments data path is created by hashing the four last digits of the its primary key. For instance, data for Experiment 57395 will be stored at {storage-directory}/experiments/73/95/57395/. This hashing approach ensures a maximum of 1000 subdirectories in any folder at a worst case scenario of 10 million experiments.
Supported Data Formats
Isabl experiments can be linked to any kind of data. Be default Isabl will match the following data types:
If you need to support more raw data formats, adding the EXTRA_RAW_DATA_FORMATS both in the api and client settings, you can extend the valid data format choices in the backend, and provide a new format file validator in the client settings or a new data importer.
ie. to support MAF format:
Tip: subclassing isabl_cli.data.LocalDataImporter and overwriting RAW_DATA_INSPECTORSmight be enough to support new data formats.
Import Data from Yaml
Isabl-CLI also supports explicit importing into a single experiment by specifying absolute file paths and metadata in a yaml file via the import-data-from-yaml command. The metadata will be added to the file_data field in an experiment's raw_data.
The two main parameters to be specified when importing are:
-fi: an argument that takes a pair of values (field, field value) to identify an experiment. For example, if you had an experiment with a
system_idofTEST_EXPERIMENT_T01, the argument would look like:--files-data: an argument that takes an absolute file path to the yaml file containing absolute file paths and metadata. For example, if you had a yaml file
/absolute/path/to/files_data.yamlwith the following contents:the argument would look like:
Full command using examples above:
View command details by running: isabl import-data-from-yaml --help
Import Reference Data and BED files
You can link reference data to assemblies and techniques. Here are a few ways of how to go about it.
Link Arbitrary Reference Data for Techniques and Assemblies
The need to register arbitrary resources for any assembly or technique (e.g. gene annotations) is also supported:
Import Assembly Reference Genome
Isabl supports the ability to track resources for assemblies and techniques. For instance, ensuring that reference FASTA files are uniformly index, named, and tracked across genome builds:
Import BED Files for Sequencing Techniques
Lastly, you can register BED files for any sequencing technique, which will be compressed, indexed, moved to the technique data directory, and registered in the database:
Imported assets are available for systematic processing by Isabl applications.
Customizing Import Logic
All registration mechanisms are configurable and can be customized by providing an alternative python sub class:
Setting Name
Default
DATA_IMPORTER
isabl_cli.data.LocalDataImporter
REFERENCE_GENOME_IMPORTER
isabl_cli.data.LocalReferenceGenomeImporter
REFERENCE_DATA_IMPORTER
isabl_cli.data.LocalReferenceDataImporter
BED_IMPORTER
isabl_cli.data.LocalBedImporter
Although only local storage is supported at the time of writing, Isabl-CLI capability can be extrapolated to cloud solutions including integration with cloud workbenches such as Arvados.
Last updated
Was this helpful?