Quick Start
⏱ tutorial time: 10 minutes
The Isabl Platform is freely available for research and academic purposes. For commercial use, a license is required. To access the source code, please get in touch with us.
Welcome to the 10 Minutes to Isabl guide! This tutorial will walk you through installation, meta data registration, data import, and automated data processing.
Also check the Retrieve Data and Register Metadata guides!
Join us on Gitter if you have questions
Submit an issue 🐛 if you are having problems with this guide
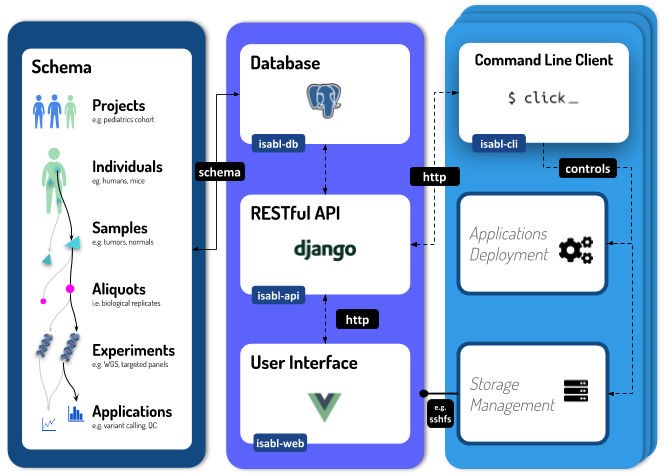
Intro to Isabl
Checkout the documentation home page for an intro to Isabl.
Prerequisites
Docker Compose for building and running the application.
Make sure your installation doesn't require sudo to run docker and docker-compose. Otherwise you will have issues running this demo. Check docker run hello-world runs without problem. If you have permissions issues, see how to run docker as-non-root user.
Demo Setup
Let's start by clone the demo:
Make sure the git submodule folders isabl_api and isabl_cli are not empty. If they are, probably the --recurse-submodules flag didn't work.
As a workaround, run:
git submodule update --recursive --init
Next, source a simple initiation profile:
If you are redoing the tutorial, we recommend to remove the demo directory and clone it again:
Also remove the Docker volume:
Installation
This installation relies on docker private images. Please make sure with the isabl team admins that you have proper access to them.
Build and run the application (this might take a few minutes):
Before running docker-compose build, specify your platform architecture if it's different from the standard intel linux/amd64, i.e. you have an Apple M1/M2:
If you are not sure, you can check your platform by running:
Now we can run the application in the background:
You can type demo-compose down to stop the application. And use demo-compose logs -f in a new session to see logs.
Create a superuser by running demo-django createsuperuser, for example using credentials: username=admin password=admin [email protected]
Now, visit http://localhost:8000/ and log in!
demo-compose, demo-django, and demo-cli are simple wrappers around docker-compose - check them out. The isabl_demo directory was bootstrapped using cookiecutter-isabl, a proud fork of cookiecutter-django! Many topics from their guide will be relevant to your project.
Create Project
Creating a project in Isabl is as simple as adding a title. You can also specify optional fields:

Configure Isabl CLI
We need to let isabl-cli know where can the API be reached, and what CLI settings we should use (if you are using demo-compose, these variables are already set):
Now we can create a client for isabl-cli:
Register Samples
Before we create samples, let's use isabl-cli to add choices for Center, Disease, Sequencing Technique, and Data Generating Platform:
New options can also be easily created using the admin site: http://localhost:8000/admin/
We will use Excel submissions to register samples through the web interface. To do so, the demo project comes with a pre-filled metadata form available at:
When prompted to allow macros, say yes. This will enable you to toggle between optional and required columns. By the way, Isabl has multiple mechanisms for metadata ingestion! Learn more here.
Now let's proceed to submit this excel form. First open the directory:
And drag the demo_submission.xlsm file into the submit samples form:

We can also review metadata at the command line:
Expand and navigate with arrow keys, press e to expand all and E to minimize. Learn more at fx documentation. Use --help to learn about other ways to visualize metadata (e.g. tsv).
For this particular demo, we wanted to create a sample tree that showcased the flexibility of Isabl's data model. Our demo individual has two samples, one normal and one tumor. The tumor sample is further divided into two biological replicates (or aliquots), with two experiments conducted on the second aliquot:

RESTful API
Although not required for this tutorial, you are welcome to checkout the RESTful API documentation at: http://localhost:8000/api/v1/ or https://isabl.github.io/redoc/.

Import Reference Data
Given that isabl-cli will move our test data, let's copy original assets into a staging directory:
Now let's import the genome:
We can also import BED files for our demo Sequencing Technique:
Check that import was successful:
By means of the --data-id flag, the command get-reference also allows you to retrieve the indexes generated during import. To get a list of available files per assembly run:
Learn more about importing data into Isabl here.
On this previous steps of the demo, we used demo-isabl to import references and bedfiles, because it is a wrapper to run isabl inside a container with samtools, bwa and tabix . These tools are needed to index these imported genomic files.
Import Experimental Data
Next step is to import data for the samples we just created:
Add --commit to complete the operation.
Retrieve imported data for the normal to see how directories are created:
The front end will also reflect that data has been imported.
Writing Applications
Isabl is a language agnostic platform and can deploy any pipeline. To get started, we will use some applications from isabl-io/apps. Precisely we will run alignment, quality control, and variant calling. Applications were previously registered in client object. Once registered, they are available in the client:
Learn more about customizing your instance with Isabl Settings.
First we'll run alignment (pass --commit to deploy):
Note that if you try to re-run the same command, Isabl will notify you that results are already available. If for some reason the analyses fail, you can force a re-run using --force.
Now we can retrieve bams from the command line:
We can also visualize aligned bams online:
Insert 2:123,028-123,995 in the locus bar, that's were our test data has reads. Learn more about default BAMs in the Writing Applications guide.

Auto-merge Analyses
Let's get some stats for our experiments with a quality control application:
This quality control application has defined logic to merge results at a project and individual level. Upon completion of analyses execution, Isabl automatically runs the auto-merge logic:

Isabl-web can render multiple types of results, in this case we will check at HTML reports. Results for our qc-data application are available at an experiment, individual, and project level. In this example we are looking at the project-level auto-merge analysis:

Applications can define any custom logic to merge analyses.
Multi-experiment Analyses
Up until now we've run applications that are linked to one experiment only. However, analyses can be related to any number of target and reference experiments. For example this implementation of Strelka uses tumor-normal pairs. Before you can run this command you will need to retrieve the system id of your experiments, let's try:
Now insert those identifiers in the following command:
You can retrieve registered results for the analysis, for instance the indels VCF:
To find out what other results are available use:
Furthermore, you can get paths for any instance in the database using get-paths:
Lastly, lets check the indels VCFs through the web portal:

Software Development Kit
To finalize the tutorial, we'll use Isabl as an SDK with ipython:
Then lets check the output directories of Strelka:
The analysis objects are Munch, in other words they are dot-dicts (like javascript):
Wrap up and Next Steps
Learn about CLI advanced configuration to customize functionality:
Isabl SettingsLearn about writing applications:
Writing ApplicationsReady for production? learn more about deployment:
Production DeploymentLast updated
Was this helpful?