Retrieving Data
🤓 The ultimate guide for data analysts using Isabl!
Introduction to Filters
Filters enable you to subset the data of your interest. For example you can use filters to retrieve all the BAM files of a given project, or get all VCFs from a given variant calling application. Filters are field-value pairs and can be used both in the Command Line and within Python. Check out this examples:
curl http://{my-isabl-instance}/api/v1/experiments?sample__identifier={the-sample-id}isabl get-outdirs -fi application.name BWA_MEM -fi status SUCCEEDEDimport isabl_cli as ii
samples = ii.get_instances('samples', individual__species="HUMAN")Note that fields can traverse the relational model. To do so concatenate the fields with __ (e.g. samples__disease__acronym=AML, or a dot in the Command Line application.name=PINDEL)
Filters Modifiers
As indicated in the previous hint, filter fields can traverse database relationships. However, all filters can be augmented using lookups:
{related_field}__{related_filter}="query"Here is a quick representation of Isabl's relational model, hence related filters:

Furthermore, all query parameters in this API support advanced lookup types:
Lookup Type
Description
Example
!
Negate any query
name!=isabel
[i]exact
Exact match
name__exact=isabl name__iexact=IsAbL
[i]contains
Value contains query
name__contains=isa name__icontains=iSa
[i]startswith
Value starts with query
name__startswith=isab name__istartswith=iSab
[i]endswith
Value starts with query
name__endswith=bl name__iendswith=bL
in
Comma separated query
name__in=isabl,besuhof
isnull
Value is null
name__isnull=true
regex
Use regex pattern
name__regex=isabl
gt
Greater than
total__gt=1
gte
Greater or equal
total__gte=1
lt
Less than
total__lt=1
lte
Less than or equal
total__lte=1
Moreover, Datetime query parameters support extra lookups:
Lookup Type
Description
Example
``
No lookup, ISO format required
created=
date
Filter by date YYYY-MM-DD.
created__date=2016-06-04 created__date__gt=2016-06-04
day
Filter by day DD
created__day=04
month
Filter by month MM
created__month=06
year
Filter by year YYYY
created__year=2016
time
Filter by time HH-MM-SS
created__time=21:00:51

To get a full description of all available filters please visit Isabl's Redoc API documentation at https://isabl.github.io/redoc/ or https://isabl.mskcc.org/api/v1 (replacing isabl.mskcc.org with your own host). Another useful way to explore the relational model is by using isabl get-metadata.
Common Filters
Here are some common and useful filters for Isabl.
Limit vs Count Limit
The filter count_limit enables you to limit the total number of instances that will be retrieved. For example to get the output directory for the first 10 successful analyses you could do:
On the other side, limit will determine how many instances should be retrieved at the same time. For example, the following command would retrieve paths to all successful analyses in batches of 10000:
Has BAM File
To get for example all experiments that have a BAM file for GRCh37 you could do:
Performance Filters
The following filters can be used to (quite dramatically) improve the performance for some queries:
Filter
Description
Usage
distinct
If you set distinct to false, the each result within the query won't be guaranteed to be unique, yet the response will be faster.
distinct=false
paginator
By activating the cursor pagination, you would be able to traverse queries results, but you won't know the total number of results.
paginator=cursor
paginator=cursor is still experimental, please report an issue if you have trouble.
Isabl Command Line Client
Filters in the command line are usually provided using the -fi or --filters flags. Relations or lookups can be provided using double underscores or dots (e.g. application.name or application__name). Here is a list of Isabl commands available to retrieve information:
Command
Description
Example
get-count
Get count of database instances given a particular query. For example, how many failed analyses are in the system?
isabl get-count analyses -fi status FAILED
get-metadata
Retrieve instances metadata in multiple formats. To limit the number of fields you are interested in use -f (i.e. --fields).
isabl get-metadata samples -fi category TUMOR -f disease
get-data
This command will retrieved the raw data linked to experiments as imported in Isabl (e.g. BAM, FASTQ, CRAM). Use --verbose to see what experiments have missing data.
isabl get-data -fi projects.pk.in 102,103
get-bams
Get the official bam registered for a given list of experiments. Use --assemblyif there are BAMs available for different versions of the genome. Use has_bam_for to filter those experiments with a registered BAM.
isabl get-bams -fi has_bam_for GRCh37
get-reference
Isabl supports the linkage of auxiliary resources to the assembly instances. By defaultget-reference gives you the path to the reference FASTA, however you can retrieve other linked resources.
isabl get-reference GRCh37
get-bed
Retrieve a BED file linked to a particular sequencing technique. By default, the targets BED file is returned, to get the baits BED use --bed-type.
isabl get-bed HEMEPACT --assembly GRCh37
get-paths
Retrieve the storage directory for any instance in the database. Use --pattern to retrieve files within those directories.
isabl get-paths projects 102
get-outdirs
This command is a short cut of isabl get-paths analyses. Learn more about retrieving results here.
isabl get-outdirs -fi name PINDEL -fi status SUCCEEDED
get-results
Retrieve analyses results produced by applications. Use --app-resultsto list all available choices for a given application.
isabl get-results -fi application.pk 1 -r command_script
Dynamically Explore Metadata
Another useful way to explore the relational model is by using isabl get-metadata:

Expand and navigate with arrow keys, press e to expand all and E to minimize. Learn more at fx documentation. Use --help to learn about other ways to visualize metadata (e.g. tsv).
Furthermore, you can limit the amount of information you are retrieving by passing the list of fields you are interested in:
Assembly Resources
By default, the command get-reference helps you retrieve the assembly reference genome.
However, by means of the --data-id flag, the command get-reference also allows you to retrieve the indexes generated during import. To get a list of available files per assembly use --resources:
Then get the one you are interested in with:
Retrieving Application Results
You can use get-outdirs within the command line to systematically explore output directories. For example:
Further more you can retrieve files within those directories by using --pattern:
Additionally, you can retrieve results directly registered by the application:
To visualize what results are available for a given application run:
You can retrieve the application primary key from the front end.
Isabl Software Development Kit
Importantly, isabl-cli can also be used as a Software Development Kit within python:
If you are using ipython, use ? to get help on a method (e.g. ii.get_instances?)
Getting Instances
To get started, we can retrieve specific instances from the database:
These instances are Munch, in other words they are dot-dicts (like javascript). So you can do both analysis["status"] or analysis.status.
A more general way to retrieve any object in the database is using get_instance:
Some examples of things you can do with these instances:
To get multiple instances you can do:
Similarly to isabl get-count , you can determine the number of available results for a given query:
Getting all Samples from an Individual
To retrieve all samples and experiments for a given individual:
You can also retrieve multiple trees:
Create, Delete, and Modify Instances
If you have permissions, you will be able to systematically alter instances in the database:
With great power, comes... yeah you know how it goes. Just be careful.
Isabl SDK Utils
Here are other useful utilities available in isabl-cli:
Method
Description
ii.api.chunks
Given a list of elements, return a list of chunks of n elements from the original list.
ii.api.api_request
Perform an authenticated request to Isabl API.
ii.api.retry_request
Retry an HTTP request multiple times with a delay between each failure.
Isabl Web
Isabl Web is a great tool to retrieve information and understand the state of affairs within the system. Simply type something in the search bar to retrieve instances across multiple schemas:

Multiple panels will be stacked horizontally as you request more information:
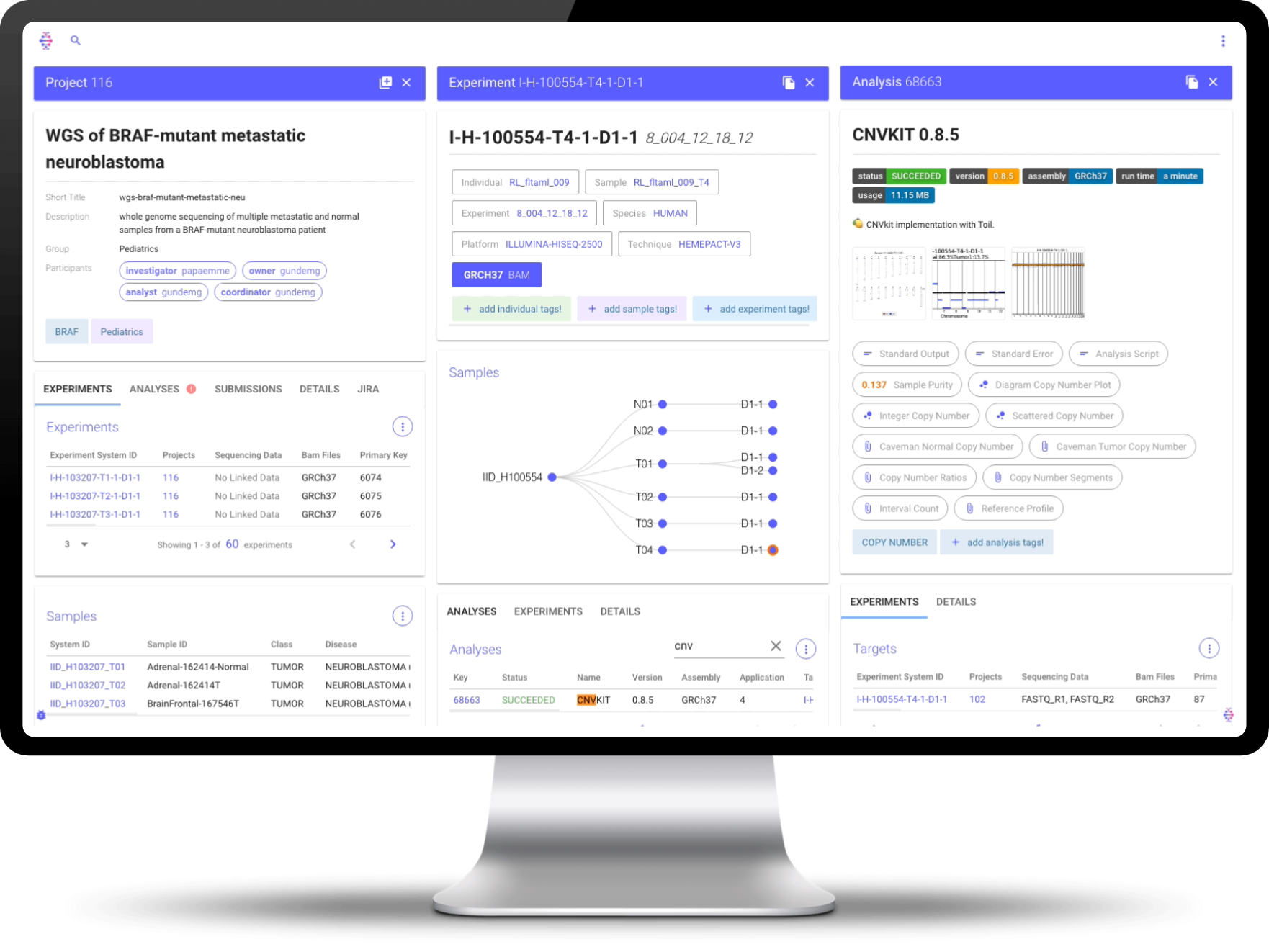
Projects Detail Panel
The projects detail panel conveys all assets and stakeholders linked to a particular project:

Live Tables are directly wired to the API and will enable you to search and filter on specific columns. For the later, simply click in the column name:

The Samples View
The samples tree panel provides access to all assets generated on a given individual.

By clicking on a given node in the tree, you can retrieve more metadata, filter out available analyses on that instance, and even get access to BAM files:

Analyses Results
We can retrieve different types of results for all analyses generated by Isabl. For example accessing a project level quality control report:

Similarly we can retrieve other types of results such as a VCF:

Last updated
Was this helpful?